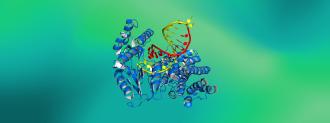
In a breakthrough decades in the making, AlphaFold, an artificial intelligence developed by London-based DeepMind, has predicted the structure of proteins with an accuracy unrivaled outside of actually dissecting them with x-rays.
The success comes in the 14th round of the Critical Assessment of Techniques for Protein Structure Prediction (CASP), a competition that tasks teams with predicting the structures of proteins based only on their amino acid sequences.
“Proteins are extremely complicated molecules, and their precise three-dimensional structure is key to the many roles they perform, for example the insulin that regulates sugar levels in our blood and the antibodies that help us fight infections,” the University of Maryland’s John Moult, co-founder and chair of CASP, said in a press release.
“Even tiny rearrangements of these vital molecules can have catastrophic effects on our health, so one of the most efficient ways to understand disease and find new treatments is to study the proteins involved.”
AlphaFold’s accuracy was high enough that CASP has called it a solution to the protein folding problem.
“This is a problem that I was beginning to think would not get solved in my lifetime,” Dame Janet Thornton of the European Bioinformatics Institute in Cambridge, UK, said in a press conference.
“Knowing these structures will really help us to understand how human beings operate and function, how we work.”
(Protein) Structure Determines Function
Proteins are fundamental to life — or, in the cases of viruses, something similar. They are made up of long strings of 20 different amino acids, which are in turn coded for in DNA.
But just because you know a protein’s genetic code doesn’t mean you can predict what it looks like. While DNA tells you a protein’s amino acid ingredients, it doesn’t tell you how all of those ingredients fit together and fold up into a 3-dimensional object.
A protein’s structure is a complex, 3D tangle of ribbons, vines, and curly fries; the amino acids fold up in very specific ways to make very specific forms. Protein folding is the only way they work; if they don’t fold correctly — or at all — the consequences can be dire.
(Take, for dramatic example, the dreaded prion, a misfolded protein that can cause other proteins to misfold, leading to a number of brain-melting diseases, most famously “mad cow” and CJD disease.)
While we know plenty of genetic codes and the amino acids they code for, being able to make the leap from those acids to what they look like as a 3D protein structure is a long, laborious, and expensive process. And the bigger and more complex the protein, the more difficult it is.
When Christian Anfinsen suggested, during his 1972 Nobel acceptance speech, that a protein’s structure should determine its function, it kicked off decades worth of work tilting at one of science’s great windmills.
There’s an incomprehensible number of possible protein structures; the Guardian‘s Ian Sample pegs the number at a googol cubed, which, if I typed it out, would be a 1 followed by 300 zeroes.
Per MIT Technology Review, labs currently determine a protein’s structure using x-ray crystallography, nuclear magnetic resonance, or cryo-electron microscopy. I won’t get into how they work here, but suffice to say, these methods can consume plenty of time and capital.
“There are tens of thousands of human proteins and many billions in other species, including bacteria and viruses, but working out the shape of just one requires expensive equipment and can take years,” Moult said.
Being able to predict the complex origami shapes of proteins based on their genetic code would open up a whole universe to scientific research.
“This really is a big deal.”
The CASP contest was inaugurated in 1994. Every two years, teams are challenged with properly predicting the structure of dozens of proteins based on their amino acid sequences. The protein structures are first worked out in a lab and then compared to the predictions of different AI or computer programs.
DeepMind had already made waves at CASP. AlphaFold had a strong showing in the 2018 edition; in 2020, it crushed it.
“This really is a big deal,” David Baker, head of the Institute for Protein Design at the University of Washington, told MIT Technology Review. (The Institute for Protein Design is behind Foldit, which makes protein folding into a game and has been competitively crowdsourcing coronavirus antiviral targets.)
“The DeepMind protein folding result is really incredible, and incredibly important,” British geneticist Adam Rutherford tweeted. But, as he noted, it’s also a hair complex.
Here’s the breakdown: CASP rates how accurate a protein structure prediction is using a measurement called the Global Distance Test (GDT). Scored from 0-100, this is essentially saying how close the structure you’ve predicted is to the amino acids’ real locations, as determined by observations with MRIs or x-ray crystallography.
A GDT score of 90 is considered pretty comparable to the current, gold-standard lab observations. This is an easy target with very small, very simple proteins, but it becomes vastly harder with bigger proteins and more complex shapes.
AlphaFold had a median score of 92.4 across all of their targets. When presented with what DeepMind’s blog characterized as the “very hardest” protein structures to predict, they scored a median of 87.0 GDT.
AlphaFold not only beat out the other computer programs and AIs entered into CASP, but it was nearly as accurate as protein structures obtained in a lab.
“This is a big deal,” Moult told Nature. “In some sense the problem is solved.”
DeepMind trained AlphaFold using a database of roughly 170,000 known protein structures from the protein data bank, as well as immense collections of protein sequences with structures unknown. Feeding all that information into AlphaFold’s deep learning neural network, DeepMind let it run for a few weeks with a “relatively modest amount” of computer horsepower.
Building on this work, AlphaFold creates highly accurate guesses of where amino acids will be in an unknown protein structure, MIT Technology Review reports.
Who Knows What the Future Folds
Understanding protein folding and protein structure could radically change how we understand any functions where proteins are involved — so, all of biology, basically.
Already, AlphaFold is helping out in the field. Andrei Lupas, an evolutionary biologist at Germany’s Max Planck Institute for Developmental Biology, has used AlphaFold to tease out a protein structure that has been flummoxing his lab for years.
“The model from group 427 (DeepMind’s CASP pseudonym) gave us our structure in half an hour, after we had spent a decade trying everything,” Lupas — who assessed high-accuracy models for CASP — told Nature.
DeepMind founder and CEO Demis Hassabis tweeted that DeepMind hopes AlphaFold “will have a big impact on disease understanding and drug discovery.”
Being able to accurately predict a protein’s structure can help researchers develop new drugs — like antibodies or antivirals that stymie SARS-CoV-2’s various proteins, including the spike — and help improve our understanding of what diseases are doing in the body.
Longer-term implications could involve helping scientists design proteins that can eat up waste, enhance biofuels, and create healthier, hardier crops.
Don’t let the trumpets drown out some of the work that’s still to be done, however.
AlphaFold hit impressive marks in ⅔ of its targets, but it showed some trouble when compared to magnetic resonance imaging, Nature reports; according to Moult, that could be a discrepancy between how the techniques turn data into a model. So far, it also has a hard time predicting protein structures in a protein complex, where several different proteins can alter each other’s folding.
DeepMind is at work on an AlphaFold paper, as well as figuring out ways to make the tool accessible to researchers.
The ultimate vision behind DeepMind has always been to build AI and then use it to help further our knowledge about the world around us by accelerating the pace of scientific discovery,” Hassabis tweeted.
“For us AlphaFold represents an exciting first proof point of that thesis.”