According to the World Health Organization, patient zero was evacuated by plane on September 12, 2008.
Initially, it began like a number of common illnesses: a spiking fever, wrenching vomiting, and diarrhea. Then came a rash, liver dysfunction, and shuddering convulsions. Her symptoms crescendoing, the safari agent was flown from the Zambian capital of Lusaka to a private medical clinic in Johannesburg, South Africa.
She died a day after admittance.
“She was cremated, and sent back to Lusaka,” says Janusz Paweska, the head of the Centre for Emerging Zoonotic and Parasitic Diseases, part of South Africa’s National Institute for Communicable Diseases (NICD). “And maybe that would be the end of the story.”
A new virus disappearing into ash.
But a little over a week after the first evacuation, another person, a paramedic, fell ill. The pathology hinted at a viral hemorrhagic fever (VHF), a vicious kind of malady caused by several viruses — like Ebola.
When these viruses replicate, they damage tiny capillaries throughout the body, causing widespread internal hemorrhaging, or bleeding, the feature that gives VHF its name.
“People are dying from hundreds of small cuts,” Paweska says.
The paramedic had treated the safari agent on the plane. It was chance that someone noticed this link in the transmission chain.
People are dying from hundreds of small cuts.
Janusz Paweska
“The admitting doctor was the same (for both patients), and he made this kind of connection between these two people,” Paweska says.
A microbiologist at the clinic reached out to the NICD, requesting help identifying the virus. The request made its way to Lucille Blumberg, then head of the NICD’s epidemiology institute; she feared an outbreak was imminent.
“I thought there was something very wrong, something going on,” Blumberg told the WHO, “and then on Thursday night we heard about the nurse who had been admitted.”
That made three.
New Virus, New Danger, New Knowledge
What had infected the agent, paramedic, and nurse was unknown — an emerging virus about to make its debut.
True to their name, these viruses emerge from the edge of the ecosystem to find bodies that have no knowledge, and therefore no defenses, against them. They can be novel, completely unknown to science, like the safari agent’s killer; they can be a highly mutated strain, like a dramatically reshuffled swine or bird flu; they can be a virus expanding its geographic range, like West Nile.
Beginning with Marburg and its more famous relative Ebola in the 1960s and 1970s, they have vaulted to various levels of infamy, an alphanumeric demonology: HIV, SARS, MERS, SARS-CoV-2, the pandemic-potential avian flus, H5N1 and H7N9.
But these are only the best known.
There are others, viruses with case counts in the single digits but which prove highly lethal. Viruses that emerge in a safari agent in Zambia and then disappear for over a decade.
These secret monsters are not singled out by the media as potential causes of a pandemic, they don’t star in war games to prevent a global health emergency, they don’t dominate the scientific conversation.
What they can do is provide us with new insights into the viruses they are related to. They can show us where mutations and recombinations are occurring, or are likely to occur, which could birth a virus capable of sparking a deadly pandemic.
New Insights into the Devils We Know
In August 2018, a brand new strain of Ebola discovered in Sierra Leone, dubbed Bombali ebolavirus, was announced to the world. By comparing Bombali with Ebola Zaire (the cause of the deadly West African Ebola epidemic), Tracey Goldstein could tease out insights about how these viruses operate.
Goldstein, associate director of the One Health Institute at UC Davis, first had to ascertain if Bombali could infect human cells using the same receptor as Ebola Zaire.
“We found that yes, yes it does do that,” Goldstein says. But importantly, the new virus infected human cells less effectively than Zaire.
Why?
When the two Ebola viruses were contrasted, differences emerged.
“When you … look at the shape of the virus, there’s a couple mutations that likely just change the shape so it’s not a perfect lock-and-key,” Goldstein says. “(Bombali is) good enough to get in, but it’s not a perfect lock-and-key.”
The location and nature of the mutations identified with the help of the new virus could prove important to understanding Ebola. They could serve as warning signs for future Ebola strains to come, a viral cobra’s hood: if you see these mutations cropping up in a new virus, beware. (The same kind of insights are being sought in the lab, in controversial gain-of-function mutation experiments.)
Identifying how Ebola enters cells can also provide targets for antiviral medications that trip up a virus’s replication cycle, a precious weapon against a new virus, if we could find one quickly — or have one ready before an outbreak happens.
The Queensland Killer’s Clues
Making the NICD virus hunter’s lives difficult was the fact that patient zero was a safari agent. Most new viruses make the leap from animals to people; the agent’s job put her in contact with any number of them. Figuring out where the virus came from — with just one spillover incident — would prove daunting.
But insights about one new virus can be drawn from others, no matter how rare. They can help us unlock the transmission cycle, identifying one more place where viral spillover may occur.
In 1999, Nipah virus emerged and infected around 300 people in Malaysia, killing 100. In the decades since, Nipah outbreaks have remained relatively small, but deadly: per the WHO, case fatality rates have ranged from 40-75%, depending on the outbreak.
Nipah also decimated the country’s pig population; millions were culled to curb the spread, an economic disaster as the pork trade was poleaxed. But the virus did not originate in pigs, and culling did not eliminate its source.
A clue to the mystery virus’s provenance laid across the Indian Ocean, thousands of miles and five years away.
First spilling over in 1994, in a suburb of Brisbane, Australia, Hendra virus has only infected seven people — but it killed four of them. Jumping from fruit bats called flying foxes to horses to humans, it’s a relative of Nipah.
Because Hendra and Nipah are so closely related, if the Australian virus came from a type of fruit bat, it stood to reason Nipah did too. Fingered by its relative, the vector for Nipah, indeed the flying foxes, was found.
With this knowledge in hand, future outbreaks of the bat-borne viruses could be better predicted, managed, and potentially even avoided.
“Not only does it give us a framework of understanding, so that we can generalize about Henipaviruses, but it also gives us a framework for looking at issues which are really, really difficult to study,” says Peter Hudson, a biology professor at Penn State.
Hendra’s had multiple bat-to-horse spillover events, so we can see why they occur and perhaps predict when, Hudson says.
“Once we have that, then we can start to understand things like Ebola, where we have very few spillover events where we have any detailed records.”
Or, in the case of a mystery virus in Zambia and South Africa, only one.
A New Virus Stalks Southern Africa
Less than a month after patient zero, NICD’s Paweska had a mounting pile of evidence — and a growing body count. Already, the safari agent and the paramedic who treated her were dead; soon after, her nurse at the clinic had died, too.
By October, the new virus had infected two more patients. One had cleaned the safari agent’s room; he died on October 6. The Bulletin of the World Health Organization lays out the terminal features, some that sound hauntingly familiar right now: respiratory distress, neurological signs, circulatory collapse.
The latest victim was another nurse, who had treated the paramedic. Her life would depend on finding the killer’s identity before it was too late.
Now that the new virus was spreading person-to-person, Paweska’s lab was tasked with figuring out the culprit. Paweska tested the blood of all the patients, except the one who had already been cremated.
“We did all of the tests which we could actually use at that time,” Paweska said. They tested for all the VHFs they knew of, to no avail. “All the PCR tests, all the molecular tests, all the serological tests, antigen detection tests — we used the whole battery of tests. They were all negative.”
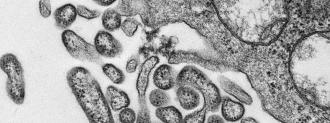
Paweska’s team needed tissue biopsies, not blood. Armed with liver samples from the deceased paramedic and nurse, he used experimental primers meant to expose Old World arenaviruses, another group of viruses that can cause VHFs. The team was able to amplify an arena-like sequence found in the samples — a sequence unlike any known to science.
“That was a surprise,” Paweska says. “It was actually quite a big surprise, because the only pathogenic arenavirus which we know of in Africa is actually Lassa fever.” Lassa virus is common in west Africa — thousands of miles away — and periodically jumps from rats to humans.
Doctors gave the nurse antiviral drugs that had worked against Lassa fever in the past. The treatment saved her life, making her the only patient to survive the outbreak.
“Dealing with a highly fatal arenavirus in southern Africa was a big surprise.”
This, too, is another lesson we can learn from a rare new virus: that where we believe viruses to live is not hard and fast, and versions and variations may be lurking where we least expect them.
The new virus now needed a name. Traditionally, a new virus was named after where it was found (like Marburg, Germany, or Hendra, Australia), a convention being abandoned for fear of stigmatization. Paweska’s Zambian colleagues in Lusaka didn’t relish the idea of “Lusaka hemorrhagic fever virus”; obviously, Johannesburg didn’t want to become the namesake either.
“After one hour of discussion, I said ‘what about Lujo?'” Paweska says. The virus came from Lusaka, and caused an outbreak in Johannesburg — Lujo.
New Virus, New Mysteries, New Answers
“We’re still very puzzled by this one-off outbreak,” Paweska says. Wherever Lujo virus was hiding, it has yet to be found — or return.
But other nasty viruses have emerged, ones that you have almost certainly never heard of — that I had never heard of. Another new VHF virus, Chapare virus, was discovered in Bolivia in 2003. Lujo arrived in 2008. Ikoma lyssavirus, a relative of rabies, was discovered in African Civets in 2012; Bombali ebolavirus in bats in 2018.
And more are sure to arise, including some that are not just variants and strains, but entirely new types of viruses we’ve never seen before.
It would be expensive (although surely cheaper than a pandemic) to genetically sequence every single new virus we find, and good luck convincing people to spend money on a virus that nobody has ever heard of, which has only infected a handful of people. But it could also unlock enormous insights, telling us where the threats are, how they operate, and how to respond — before they spill over the species boundary.
“Can it use the human receptor?” Goldstein says. “How well do they replicate in its host cell versus human cells? Can they modulate the immune system?”
As more zoonotic diseases arise, spurred on by viral evolution and changing climates, the lessons that can be learned from any virus — no matter how small the outbreak — may prove crucial. The secret monsters that do not make the news are heralds, warnings of what could come, proof of all that we don’t know.
Like, where is Lujo now? And when is it coming back?